International Research Projects:
- TRANS GLIOMA: Promotion of a Joint Translational Platform to Identify Innovative Therapies to Treat Glioblastoma (EC Interreg V-A Italy-Slovenia Transborder Project).
- 4D-HEALING: Data-Driven Drug Discovery for Wound Healing (ERA CoSysMed Project).
- Molecular basis of keratinopathies (CELSA Project).
- Generation of nanobodies against immunomodulating checkpoint receptors in glioblastoma tumor cells (CELSA Project).
National Research Projects:
- Identification of novel glioblastoma bio-markers for non-invasive liquid biopsy (Post-doctoral Research Project).
- Zinc finger 714 as a potential biomarker of suicidal behaviour (Post-doctoral Research Project).
- Development of anti-FREM2 nanobody and its use for targeting glioblastoma cells (Post-doctoral Research Project).
- Early detection of Alzehimer’s disease with methylation status of candidate genes in liquid biopsies cfDNA (Basic Research Project).
- Genetic basis of erythrocytosis in Slovenia (Applied Research Project).
######################################################################################################
International Research Projects
TRANS GLIOMA: Promotion of a Joint Translational Platform to Identify Innovative Therapies to Treat Glioblastoma
______________________________________________________________________________________________________________________________________________
Project Coordinator: Prof. Radovan Komel, PhD
______________________________________________________________________________________________________________________________________________
| Acronym: TRANS-GLIOMA Duration: 1.9.2017 – 30.11.2020 |  |
 |
The EC Interreg V-A Italy Slovenia Transborder Research Project is financed by the European Funds for Regional Development.
Summary
The aim is to increase the cooperation between the key actors in the field of biomedicine (research institutes, hospitals, universities and enterprises), to promote the transfer of knowledge and innovative biomedical techniques in the field of brain cancer (glioblastoma) and improving the competitiveness of research in oncology.
The project is upgrading the results of the previous GLIOMA project (2007-2013), validating the prognostic values of detected biomarkers in a large number of cases and their use in finding new therapeutic options. Strengthening the cross-border network of partners will increase competitiveness in research and treatment.
Expected results are: development of open-source code software, designed to analyze the data of the joint bio-bank of project partners (called GLIOBANK), identification of a new personalized pharmacological targets for patients, close collaboration of 3 research organizations, 2 hospitals and 1 enterprise and others involved.
This set up collaboration will benefit for researchers, medical doctors, students, spin-offs and start-ups and of course patients. The approach is based on the presence of a critical mass of centers of excellence (universities, research institutions and enterprises) and on the transfer of the research results into the market. In order to achieve this cross-border approach, specific but different competences are needed. The project is innovative with introduction of “precision medicine” into glioma field, with identification of the genomic profile of the tumor microenvironment and patient tumor stem cells. It will lead into using new, specific medicinal products which are specific for certain subtype of the disease on the individual level, increasing the possibility of using a new way of treatment for currently incurable disease.
Project Partners
• PP1 Leading Partner: University of Ljubljana (IL) – Faculty of Medicine
• PP2 Azienda Sanitaria Universitaria Integrata di Udine (ASUIUS), University of Udine
• PP3 Elettra Sincrotrone Trieste S. C. p. A.
• PP4 National Institute of Biology (NIB)
• PP5 Biosistemika d.o.o.
• PP6 Azienda ULSS 3 SERENISSIMA
Associated Partners of the Project
• Chamber of Commerce and Industry of Slovenia
• Slovenian Innovation Hub European Economic Interest Grouping
• Centre of Excellence for Biosensors, Instrumentation and Process Control (COBIK)
• SiNAPSA, Slovenian Society for Neuroscience
• Direzione centrale salute, integrazione socio sanitaria, politiche sociali e famiglia “Area servizi assistenza ospedaliera – Regione Autonoma Friuli Venezia Giulia
Value of the project: 1.302.252,49 €
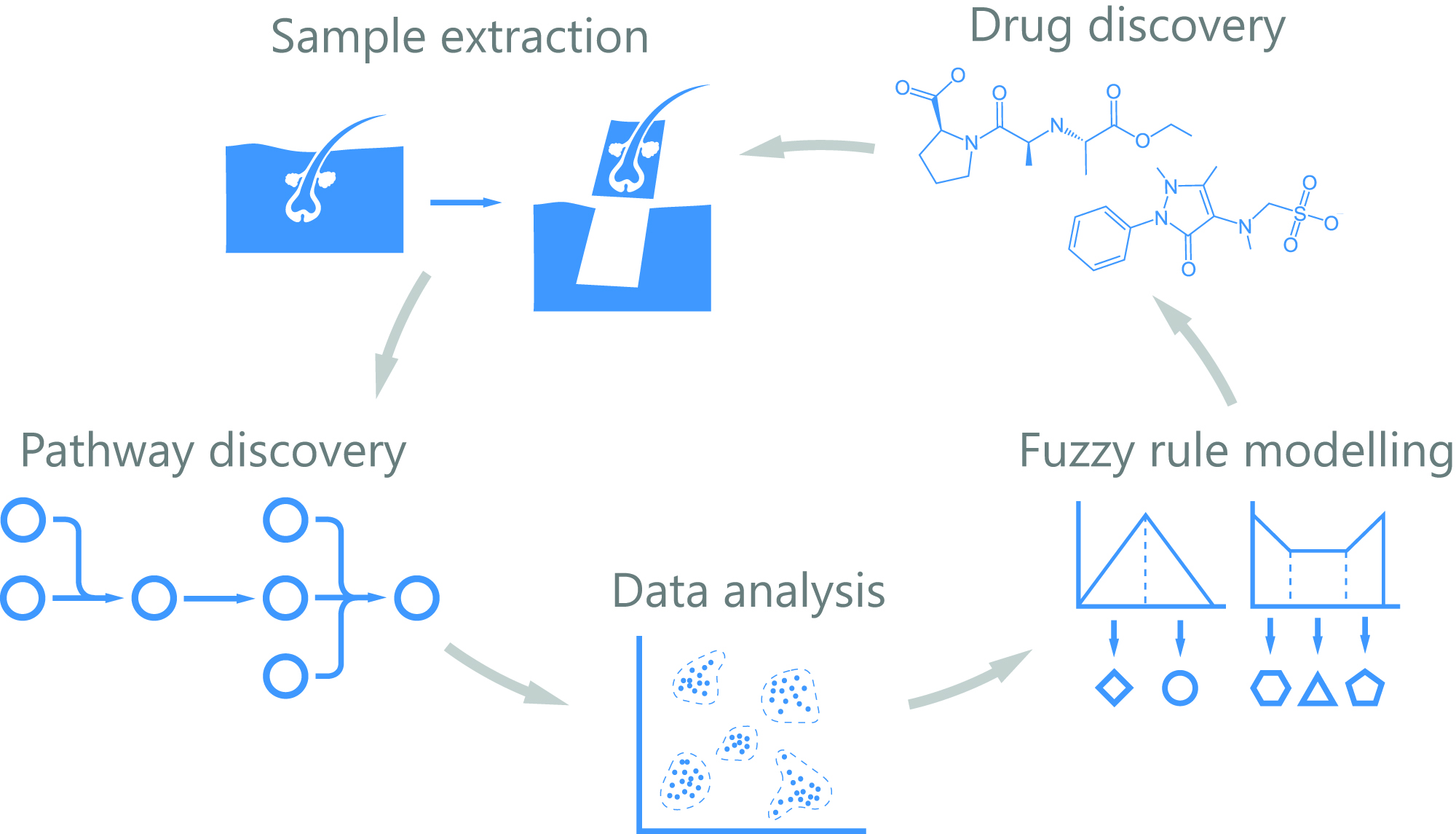
Data-Driven Drug Discovery for Wound Healing
_____________________________________________________________________________________________________________________________________________ Slovenian Project Leader: Prof. Radovan Komel, PhD Slovenian PI: Mirjana Liović, PhD _____________________________________________________________________________________________________________________________________________

Acronym: 4D-HEALING Duration: 1. 6. 2018 – 31. 5. 2021
The ERACoSysMed project has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No 643271.
Summary
The healing of wounds is a complex, multifactorial process that remains incompletely understood. Health providers in Europe face an increasing number of costly chronic wounds which profoundly affect patients’ quality of life. Wound healing is highly a dynamic and heterogeneous process, which impedes the dissection of the contribution of the different cell lineages by bulk transcriptomics analysis.
In addition, a static snapshot by single cell analyses might be uninformative. 4D-HEALING will map the four spatiotemporal dimensions of human wound healing in an unprecedented way: the transcriptome of thousands of cells from an in vivo monitored human wound will be analysed by RNAseq at the single cell level. We will model the transcriptional dynamics of cell lineages involved in all phases of healing, from injury to resolution. To gather positional information we propose a novel computational tool to predict “cell docking”, based on the probability of cell-to-cell interactions and produce a neighbouring map that will be validated by annotating positional information of cells (multiplexed hybridization approach).
The emerging knowledge will provide a 4D map of the cell types involved in human wound healing allowing us to create a dynamic mathematical model, to disentangle the crosstalk among cell signalling and interactions, transitional states and position during the healing process. These models integrated together with FDA-approved drug databases will permit informed choices on molecular candidates that can be pharmacologically modulated. Candidate compounds will be tested in ex vivo human skin organ cultures mimicking acute and chronic wound environments. The resulting leads will undergo standard drug development programs.
The project will deliver data-driven, comprehensive understanding of the human wound healing process and in vitro proof of concept of a novel topical therapy to enhance chronic wound healing. Understanding wound healing will shed light to regenerative medicine.
Project Partners
• Biodonostia Health Research Institute, Spain – Project Coordinator
• Monasterium Laboratory Skin & Hair Research Solutions GmbH, Germany
• University of Ljubljana, MCMB, Faculty of Medicine, Slovenia
• Mathematical Institute, Slovak Academy of Sciences, Slovakia
• Medical University of Vienna, Austria
• DEBRA Austria, Austria
• University of Manchester, United Kingdom
• European Bioinformatics Institute, Sanger Institute, United Kingdom
Link to the 4D-HEALING web site: 4D-HEALING
Molecular basis of keratinopathies
______________________________________________________________________________________________________________________________________________ Project Coordinator: Sergei Strelkov, PhD Slovenian PI: Mirjana Liović, PhD ______________________________________________________________________________________________________________________________________________ 
| Duration: 1. 1. 2019 – 31. 12. 2019 | |
The CELSA project has received funding from the Central Europe Leuven Strategic Alliance.
Summary
A range of congenital skin blistering diseases, including the notoriously known epidermolysis bullosa simplex (EBS), are linked to mutations in keratins. These proteins form the intermediate filaments present in the cytoplasm of epithelial cells. Mutations in keratins typically cause skin fragility in a dominant way, causing constant suffering of the affected individuals (“Butterfly Children”). At this moment there is no effective treatment for these genetic diseases. This proposal originates from three research laboratories in Leuven, Ljubljana and Prague with pronounced complementarity in technical expertise. Two of us have extensive track record in the intermediate filament research already while the third one excels in the chemical crosslinking technique which is central to this proposal.
Our goal is to investigate the molecular basis of skin fragility caused by keratin mutations, which is an indispensable first step towards a rational design of future therapies. We will investigate the effect of mutations linked to severe EBS phenotypes, such as keratin K5 mutation E475G and keratin K14 mutation R125P, on the filament assembly in vitro. These keratins form heterodimers when mixed together, which then associate to tetramers and eventually produce long mature filaments. After in vitro co-assembly, chemical crosslinking coupled to mass-spectrometric analysis will be used to structurally analyse the assembly process, towards delineating a specific point at which the assembly is affected by mutations. In parallel, electron microscopy studies using both negative staining and unstained samples in vitreous ice will be performed. In addition, crystallization of truncated heterotetramers will be attempted. Next, we will study the effect of the mutants in cell culture, to be obtained using the induced pluripotent stem (iPS) cell technology. Through epifluorescent microscopy on EGFP-labelled keratins we will document changes in the cytoskeleton caused by these mutations. We will also examine the changes in the mechanical properties of cells by using optical tweezers and live cell imaging. As the result of this multipronged in vitro and cell culture study, we expect to fully characterize the effect of several mutations on keratin filament structure and properties. This should serve as a paradigm towards a mechanistical description of keratinopathies, which is an indispensable step towards developing a successful therapy in the future.
Project Partners
• Sergei Strelkov, KU Leuven, Department of Pharmaceutical and Pharmacological Sciences, Biocrystallography, Belgium
• Mirjana Liović, UL, Faculty of Medicine, Medical Centre for Molecular Biology, Institute of Biochemistry, Slovenia
• Petr Novak, CUNI, Faculty of Science, Laboratory of Biomolecular Structure and Function, Czech Republic
Link to CELSA web site: CELSA-2018
*******************************************************************************************************************************************************************************
Generation of nanobodies against immunomodulating checkpoint receptors in glioblastoma tumor cells
______________________________________________________________________________________________________________________________________________ Project Coordinator: Frederik De Smet, PhD Slovenian PI: Ivana Jovčevska, PhD ______________________________________________________________________________________________________________________________________________ 
| Duration: 01.10.2020 – 30.09.2022 | |
The CELSA project (No. 3M200315) has received funding from the Central Europe Leuven Strategic Alliance.
Summary
Glioblastoma is the most aggressive intrinsic brain tumor in adults. In spite of maximal therapy consisting of surgery, radiotherapy and chemotherapy, glioblastoma remains largely incurable with a median overall survival of less than 15 months. Preclinical and early clinical research in immunotherapy for brain tumors has a track record of almost 20 years and indicated that an immune reaction against the tumor in the brain can be elicited and that several types of immunotherapy are feasible, safe and can result in anecdotical cases of long-term surviving patients in a good quality of life. Nevertheless, immunotherapy for brain cancer has not entered the arena of standard clinical practice unlike many other cancer types. One of the main reasons is that virtually all large immunotherapy trials for glioblastoma have ignored the brain specific constitution of the immune system and have focused on the same immune checkpoint targets as for peripheral tumors. However, current expression data suggest that other checkpoint regulators apart from PD-L1 are more abundantly expressed by GBM tumor cells, including members of the B7 family, and should therefore be considered as well. For these alternative targets, reagents to study their functional role are still largely missing however.
In this project, we therefore aim to generate nanobodies against the 4 most abundantly expressed immunomodulating checkpoint receptors in glioblastoma tumor cells. This will be done by screening and expanding a pre-existing library of nanobodies that was generated following the immunization with a patient-derived glioblastoma cell line. Panning and selection will be done using the extracellular domains coupled to Fc-fragments, following binders will be selected using high and low expressing patientderived glioblastoma cell lines and negative controls (either other tumor cells or Crispr inactivated cells). Finally, using competition assays, we will select those nanobodies that are able to prevent the binding of the endogenous ligand to each receptor.
This project will generate the foundation for novel reagents that can be used to assess the importance of the variety of checkpoint molecules in the modulation of the immune responses in the brain. Once nanobodies have been identified, they will be assessed in preclinical mouse models for their potential activities.
Project Partners
• Frederik De Smet, KU Leuven, Translational Cell & Tissue Research, Laboratory for Precision Cancer Medicine, Belgium
• Ivana Jovčevska, UL, Faculty of Medicine, Medical Centre for Molecular Biology, Institute of Biochemistry, Slovenia
Link to CELSA web site: CELSA-2020; CELSA
##############################################################################################################
National Research Projects
Identification of novel glioblastoma bio-markers for non-invasive liquid biopsy
______________________________________________________________________________________________________________________________________________ Project Leader: Neja Šamec, PhD ______________________________________________________________________________________________________________________________________________ 
| Duration: 1. 9. 2020 – 30. 8. 2022 Range: 1.0 FTE |  |
The post-doctoral basic research project Z3-, in the annual volume of 1.0FTE, price category C, is financed by the Slovenian Research Agency (ARRS) from the state budget.
Summary
Glioblastoma is the most common and lethal form of brain tumor. Due to appearance of non-specific symptoms and aggressiveness of the tumor, early disease diagnosis is highly unlikely. Current diagnostics involves detection of tumor with magnetic resonance imaging (MRI), which does not differentiate between actual tumor progression and necrotic tissue after surgery and has limitations in terms of resolution about 2-3 mm. Therefore the tumor is present in the brain before we can even detect it. Moreover sampling of tumor tissue does not capture the entire tumor heterogeneity. In this project we will determine expression changes of circulating blood biomarkers from plasma of patients with 60 different grades of glioma compared with 40 healthy individuals. The selected biomarkers, ALYREF, DPYSL2, FREM2, TUFM, TRIM28, VIM, CRMP1, SPRY1 and NAP1L1 were, in our previous research, identified as differentially expressed in glioma tissue samples compared to lower grade gliomas and reference non-tumor brain samples. The first step of the project will be optimization of protocols for isolation of extracellular vesicles and RNA from blood. We will determine the more suitable protocol for isolation of RNA from blood, with higher yield and better quality. After the protocol will be optimized, we will proceed with isolation of total RNA, miRNA and proteins from 60 glioma patients. Next, the determination and validation on proteomic and transcriptomic levels of over expressed glioma biomarkers ALYREF, DPYSL2, FREM2, TUFM, TRIM28, VIM, CRMP1, SPRY1 andNAP1L1 will follow. RNA expression levels will be determined with digital droplet PCR due to the high sensitivity of the method which is crucial for biological samples where the amounts of the RNA of interest are limited. At the proteomic level, we will study the expression of ALYREF, DPYSL2, FREM2, TUFM, TRIM28, VIM, CRMP1, SPRY1 and NAP1L1 proteins with western blot. For 8 glioblastoma patients who will be enrolled in chemotherapy treatment with TMZ, we will follow the expression of the aforementioned biomarkers on proteomic and transcriptomic level to determine if they can be used as biomarkers for monitoring the disease. In the second year of the running project we will examine the effect of micro (mi) RNA and long non-coding (lnc) RNA on changes in expression of circulating blood biomarkers in 8 patients with glioblastoma compared to 8 healthy individuals. Evaluation of miRNA and lncRNA with significantly different expression in glioblastoma samples against samples isolated from plasma originating from healthy individuals will be performed and construction of the mRNA-lncRNA-miRNA co-expression network will be determined. With this research we will provide accessible biomarkers with non-invasive liquid biopsy for early glioblastoma detection and future successful treatment monitoring.
This research is financed by the Slovenian Research Agency. The experimental work will be performed at the MCMB, Institute of Biochemistry, Faculty of Medicine UL.
Project group structure: SICRIS
Bibliographic data and other project information: Z3-2469
*******************************************************************************************************************************************************************************
Zinc finger 714 as a potential biomarker of suicidal behaviour
______________________________________________________________________________________________________________________________________________ Project Leader: Katarina Kouter, PhD ______________________________________________________________________________________________________________________________________________ 
| Duration: 1. 9. 2019 – 30. 8. 2022 Range: 1.0 FTE |  |
The post-doctoral basic research project Z3-, in the annual volume of 1.0FTE, price category C, is financed by the Slovenian Research Agency (ARRS) from the state budget.
Summary
Suicidal behaviour is a grave public health problem with nearly one million lives lost each year. Despite the decline in suicide rate, Slovenia is still ranked as a country with one of the highest rates of suicide. In previous DNA methylation studies on Slovenian suicide victims we identified zinc finger protein 714 (ZNF714) as a potential biomarker. Besides being differentially methylated in suicide victims it showed remarkable similarity in the methylation pattern between four different brain regions and blood. ZNF714 and its DNA methylation pattern could therefore be used as a blood marker of suicidal behaviour. Little is known about ZNF714 and its role in cellular processes. Gene ontology terms associate it with DNA binding and transcriptional regulation. Based on the knowledge of the protein domain structure, ZNF714 could be implicated in gene silencing via mechanism of posttranslational histone modification. However, the genes to which ZNF714 would bind in the case of transcriptional regulation are unknown.
Therefore, the aim of this project is the molecular investigation of ZNF714 on a genetic, epigenetic and protein level using tissue samples of suicide victims and control group. Proposed project will be carried out through 5 main work packages, which include chromatin immunoprecipitation sequencing (ChIP-seq), protein expression level analysis and reanalysis of existing sequencing data. Results, collected through work packages, will characterise the ZNF714 protein binding motif and genes, on which it binds; and analyse protein expression level of ZNF714 and its potential protein binding partners, involved in transcriptional regulation. Finally, the existing data will be re-analysed, focusing on the ZNF714 gene and the genomic regions identified with ChIP-seq (regions of the genome on which the ZNF714 protein binds). In addition to DNA methylation we will analyse the presence of single nucleotide polymorphisms (SNPs) and atypical non-CpG methylation (potentially methylated cytosines are not followed by guanine as usual but by some other base). The purpose of this project is therefore to gain a better understanding of ZNF714, which will allow possible clinical collaboration and future use of biomarkers in psychiatry.
Being a country with a high rate of suicide, our work makes an important contribution to the growing knowledge of suicidality research. There is an unmet need for sensitive and specific biomarkers in the field of psychiatry. A more detailed characterisation of ZNF714 could help us to achieve a better understanding of suicidal behaviour aetiology. Additional clinical studies are needed, but our project can present a step towards the use of ZNF714 as a biomarker. An important aspect of our work also consist of working towards destigmatisation of mental disorders and suicidal behaviour, which is achieved by participating in conferences, meetings and discussions aimed at both professional and lay public.
This research is financed by the Slovenian Research Agency. The experimental work will be performed at the MCMB, Institute of Biochemistry, Faculty of Medicine UL.
Project group structure: SICRIS
Bibliographic data and other project information: Z3-2653
*******************************************************************************************************************************************************************************
Development of anti-FREM2 nanobody and its use for targeting glioblastoma cells
______________________________________________________________________________________________________________________________________________ Project Leader: Ivana Jovčevska, PhD ______________________________________________________________________________________________________________________________________________ 
| Duration: 1. 7. 2019 – 30. 6. 2021 Range: 1.0 FTE |  |
The post-doctoral basic research project Z3-1869, in the annual volume of 1.0FTE, price category C, is financed by the Slovenian Research Agency (ARRS) from the state budget.
Summary
Glioblastoma multiforme is the most frequent and malignant brain cancer, which is typical for Caucasian men in an advanced age (Agnihotri S., et al. PMID: 23224339). Because of the unspecific symptoms (headache, confusion, and memory loss) early diagnosis is highly unlikely. Standard of care for glioblastoma patients comprises maximal surgical resection, followed by radiation and chemotherapy, but it is not curative. However, despite the efforts of the biomedical research patient survival remains to be 12 to 18 months after diagnosis (Veliz I., et al. PMID: 25705639, Agrawal NS., et al. PMID: 25346943, Bloch O., et al. PMID: 28193626, van Woensel M., et al. PMID: 24202332, Gruber ML., et al. PMID: 14758131, Ma C., et al. PMID: 28003765). The three main problems in glioblastoma management are fast tumor growth, intracranial location and late diagnosis. With the current clinical treatment glioblastoma remains a terminal illness with 2- and 5-year survival rates being 26% and 5%, respectively (Argawal NS., et al. PMID: 25346943, Veliz I., et al. PMID: 25705639). The fast progression and high mortality rate of glioblastoma only emphasize the urge for developing novel treatment methods. Despite the growing number of publications on “cancer bio-markers”, there is no single molecule that can be used as a universal marker for the disease. Therapeutic methods should be developed in the direction of targeting a specific subset of cells or a cellular characteristic that is crucial for tumor development. For minimizing damage on adjacent tissue, identification of specific glioblastoma bio-markers is necessary for development of targeted therapeutic approaches which will eventually lead to improvement in patient care and life expectancy.
The aim of this research is obtaining and functional characterization of a nanobody targeted against the protein of interest – FREM2. In our previous study (Vidak M., et al. PMID: 29734672) using an in silico approach we identified FREM2 as a potential glioblastoma bio-marker. FREM2 gene and protein expression was examined using glioblastoma cell lines and astrocytes. An interesting finding was surface expression of FREM2 in glioblastoma cells which was not observed in the case of the reference cell line. Furthermore, to assess potential use of FREM2 for clinical purposes, we examined gene and protein expression levels in human glioma and reference tissue samples, and performed large-scale in silico analysis using data available from The Cancer Genome Atlas Project (Jovčevska I., et al. PMID: 31357584). Our findings show that FREM2 is positively associated with survival in glioblastoma IDH-WT and negatively associated with survival in low grade glioma. Due to its surface expression in glioblastoma cells FREM2 is an attractive target for development of targeting mechanisms. Obtained anti-FREM2 nanobody can be further developed into a biological nano-drug if proven it shows specificity for toxicity on glioblastoma cells over non-malignant astrocytes. The obtained nanobody can also be used as a carrier i.e. to transport a cytotoxic agent to a specific cell type. Nanobodies have already proven useful in the selection of disease specific bio-markers (Jovčevska I., et al. PMID: 25419715, Jovčevska I., et al. PMID: 28498803, Šamec N., et al. PMID: 29707108). Nanobodies are the smallest naturally occurring antigen-binding fragments with unique antigen binding specificity (Muyldermans S. PMID: 23495938). They possess a number of advantages like small size, stability against non-physiological pH and extreme temperatures, water soluble and economically produced, which make them suitable for research purposes. With this work we expect to obtain a nanobody with high specificity and affinity towards its antigen – FREM2. Such nanobody can be used for selective targeting of glioblastoma cells with minimal damage to adjacent cells.
This research is financed by the Slovenian Research Agency. The experimental work will be performed at the MCMB, Institute of Biochemistry, Faculty of Medicine UL and in cooperation with the Jožef Stefan Institute in Ljubljana.
Bibliographic data and other project information: Z3-1869
*******************************************************************************************************************************************************************************
Early detection of Alzehimer’s disease with methylation status of candidate genes in liquid biopsies cfDNA
____________________________________________________________________________________________________________________________________________ Project Leader: Alja Videtič Paska, PhD ____________________________________________________________________________________________________________________________________________ 
| Duration: 1. 7. 2019 – 30. 6. 2021 Range: 0.75 FTE |  |
The basic research project J3-9293, in the annual volume of 0.75 FTE, price category C, is financed by the Slovenian Research Agency (ARRS) from the state budget.
Summary
Alzheimer’s disease (AD) is a slow, irreversible, but progressive, complex and multifactorial neurodegenerative disorder, which represents the most common cause of dementia in older population, and a major health problem worldwide. Due to the accelerated aging of human population, it is assumed that the number of AD patients will quadruple by the year 2050. The risk of developing AD significantly increases after 65 years of age, and it reaches up to 31% for individuals beyond age 85. The treatment of AD was not significantly changed or improved in the last decade: it includes acetylcholine esterase inhibitors donepezil, galantamine and rivastigmine, and NMDA receptor antagonist memantine. Although a lot of new drugs with novel mechanism of action were effective in animal models, only a few of them showed clinical efficacy in improving cognitive decline. Main risk factors for AD are older age, genetic predisposition, gender, cardiovascular factors and presence of the mild cognitive impairment (MCI). MCI is characterized by the slight cognitive changes and disruptions that occur in mild changes in memory, the ability to think and to remember. MCI might lead to AD, since a great percent of subjects with MCI (50 % – 65 %) later develop some form of dementia, especially AD. At present, clinical diagnosis of (probable) Alzheimer’s disease is established thorough a combination of clinical symptoms, cognitive screening tests, detailed neuropsychological testing and imaging techniques. Tests based on molecular-genetic biology analysis are only entering the routine clinical practice, but are as are other clinical tests, relevant only after the disease has already made considerable progress.
In our study we will use most contemporary methods (next generation sequencing, droplet digital PCR) to determine methylation status and single nucleotide polymorphisms of AD candidate genes, catechol-o-methyl transferase and brain-derived neurotrophic factor, in blood-based liquid biopsy samples and cell-free DNA (cfDNA) from plasma in clinically well-defined AD patients and subjects with MCI.
The introduction of additional epigenetic markers in a set of clinical tests that are currently in use in the diagnosis of AD would enable a more accurate diagnosis in the early stages of the disease, reducing the number of false-positive and false-negative diagnoses. Determining the differences of white blood cells methylation status and methylation cfDNA as a pool of DNA from distant tissues could confirm existence of the so-called ‘mirror effect’ where the peripheral markers reflect the status of unobtainable tissue for molecular studies. The identification of integrative biomarkers would mean a large step forward for diagnostic procedures of Alzheimer’s disease and for the prediction of its progression, probably also for the pre-clinical stages of the disorder like MCI, and through a translation to clinical practice.
The purpose of the study is to contribute to better understanding of epigenetic changes and their role in the underlying AD pathophysiology, and behavioural and psychological symptoms of dementia and cognitive decay. The early diagnostics would importantly contribute also to prevention and importantly affect life of AD patients and also their families. More rational pharmacotherapy could therefore ease the burden on the health funds.
Project group structure: SICRIS
Bibliographic data and other project information: J3-9293
*******************************************************************************************************************************************************************************
Genetic basis of erythrocytosis in Slovenia
______________________________________________________________________________________________________________________________________________ Project Leader: Nataša Debeljak, PhD ______________________________________________________________________________________________________________________________________________ 
| Duration: 1. 7. 2018 – 30. 6. 2021 Range: 0.75 FTE |  |
The applied research project L3-9279, in the annual volume of 0.75 FTE, price category C, is financed by the Slovenian Research Agency (ARRS) from the state budget.
Summary
Within the project Genetic basis of erythrocytosis in Slovenia (GenEry) we aim to addresses current unmet diagnostic needs for a rare haematological condition, congenital erythrocytosis. More than 60% of patients in EU are left underdiagnosed, moreover diagnosis in Slovenia is not yet established.
Erythrocytosis is a haematological condition with increased mass of erythrocytes and consequently increased haematocrit and haemoglobin. It is caused by inherited or acquired mutations or as a compensatory mechanism in some chronic diseases (heart, lung, kidney, etc). Clinical signs are often overlooked until complications; e.g. thrombosis that could result in mortality. Therefore rapid and accurate diagnosis is of high importance.
The genetic cause of erythrocytosis may be polycythemia vera or congenital (familial) erythrocytosis. Polycythemia vera is the most common acquired erythrocytosis and JAK2 mutations are routinely diagnosed, also in Slovenia. Congenital erythrocytosis however has heterogeneous genetic background, making the diagnosis very difficult. Reasons can be mutation in the genes involved in detection of oxygen levels in the blood (EPO, EPOR, VHL, EGLN1, EPAS1) or regulation of haemoglobin affinity to oxygen (HBA1, HBA2, HBB, BPGM, PKLR). Within the European Congenital Erythrocytosis Consortium only 7 out of 21 genes currently associated with the congenital disease are genetically diagnosed using classical but time consuming Sanger sequencing. Due to poor diagnosis treatment is not well defined and medical doctors do not share a common opinion regarding best treatment selection (low dose aspirin, venesection, etc.).
Within the project we will apply holistic, NGS diagnostic solutions in aim to update diagnostic algorithm and focus on discovering new disease mechanism. Development of NGS for the diagnosis of congenital erythrocytosis will enable faster and cheaper analysis of patients and simultaneous analysis of all target regions/genes of interest in one test. Project is imply 5 working packages focusing on diagnostic as well as clinical aspect of disease. Within WP1 we will evaluate etiological data of disease in Slovenian population and update current diagnostic algorithm. WP2 will focus on development of classical Sanger sequencing as well as new NGS diagnostic approach. We will analyze genes previously associated with the disease as well as new genes, including exonic, intronic and regulatory regions. WP3 is designed to understand functional impact and molecular mechanism of newly discovered genetic variants, subsequently in updated disease model. Within the WP4 we will design and validate NGS panel for research of idiopathic erythrocytosis. Furthermore, we will plan to develop additional simple and quick genetic tests for verification of most commonly discovered genetic variants. The focus of WP5 is management and disseminatioion.
Improved, quick and cheaper diagnosis developed within our project will unable to better understand disease mechanism and consequently patient stratification. Thereafter the best treatment practice to each patient subgroup will be possible (personalized medicine).
Project group structure: SICRIS
Bibliographic data and other project information: L3-9279
######################################################################################################
Past Research Projects
J3―7132: Epigenetic factors and gene expression in suicidal behaviour (1.1.2016―31.12.2018; A. Videtič Paska) [SICRIS]
BI-MK/17-18-003 Bilateral project Slovenia-Macedonia: Analysis of forensic and ancestry informative Y chromosome SNPs (A. Videtič Paska; 2017-2018).
J3-7320 Influence of different neuronavigation procedures on treatment efficacy of repetitive transcranial magnetic stimulation in depression; basic research project, [P. Pregelj (University Psychiatric Clinic Ljubljana): Project Leader; A. Videtič Paska: Leader of the research group at the Faculty of Medicine; 1.1.2016 – 31.12.2018] [SICRIS]
J3―5504: The role of polymorphisms in segregation genes in cancer (1.8.2013―31.7.2016; R. Komel) [SICRIS]
INTERREG EC Project 2011, Ref. No. 42: Identification of novel biomarkers to brain tumors – glioma, for diagnosis and as new therapeutic targets (Acronym: GLIOMA) [R. Komel, Project Leader/Coordinator for the University of Ljubljana; T. Lah Turnšek, NIB, General Project Leader; 1/11/2011 ― 30/04/2015] [GLIOMA]
J3―3617: Regulation of gene expression and cell signaling in epidermolysis bullosa simplex: a hereditary skin fragility disorder with many faces (1.5.2010―30.4.2013; M. Liović) [SICRIS]
J4―2212: Fungal cytochromes P450, involved in detoxification of plant defence compounds, as targets of novel antifungal drugs (1.5.2009―30.4.2012; N. Kraševec & R. Komel) [SICRIS]
J3―2274: Keratins and reduction of cellular adhesion: consequences for the fragility of the epithelium in chronic bowel inflammation (1.5.2009―30.4.2012; M. Liovič) [SICRIS]
J3―0124: The role of recombinant human erythropoietin on gene expression and signal transduction in breast cancer (1.2.2008―30.1.2011; N. Debeljak) [SICRIS]
Z3―0072: Polymorphisms and mutations in chromosomal segregation genes in Slovenian patients with gastric cancer (1.2.2008―30.1.2010; P. Hudler) [SICRIS]
J1―9740: Development of protein microarray for research on gastric cancer proteome (1.1.2007―31.12.2009; R. Komel) [SICRIS]
Z3-9403: Molecular mechanism of action of the recombinant human erythropoietin on cancerous cells, post-doc research project (1/1/2007 – 31/12/2008; S. Berne) [SICRIS]
ERDF Project – Centre of Excelence (CoE) »Biotechnology with Pharmacy«; Consortium Coordinator: R. Komel, MCMB MF UL (20 SLO partners institutions/laboratories; 2004 – 2007) [ CoE BF]
Z1―7562: Cross-talk between cell cycle regulation and cholesterol & drug metabolism in the mammalian liver (1.9.2005―31.8.2007; R. Komel) [SICRIS]
J3―6132: Dynamics of cytoskeleton of keratin intermediate filaments and their role in the development of the disease phenotype (1.2.2004―30.1.2007; M. Liović) [SICRIS]
PICS-3294 Project »Characterisation of proteome markers in cancer cell lines and tissues using surface plasmon resonance imagery on chip« [Joint Research Project of the International Agreements of Slovenian Government (Ministry for High Education, Science and Technology) with French Government (CNRS, France)]. R. Komel: SLO Project Leader (D.Pompon, CNRS, Gif-sur-Yvette, France: French Project Leader), MF UL: 2004 ― 2006. [PICS]
L3―5016: Transcription of anorexia candidate genes and an attempt of elaborating DNA microarray (1.1.2003―31.12.2005; R. Komel) [SICRIS]
L4―4353: Steroid 11beta Hydroxylase from the filamentous fungus Cocliobolus lunatus (1.7.2002―30.6.2005; R. Komel) [SICRIS]
Z1―4286: Erithropoietin (Epo) signal transduction pathway and mechanism of singal transduction to c-myc (1.7.2002―30.6.2004; R. Komel) [SICRIS]
Z3―3310: Keratin cytoskeleton dynamics and disease phenotype development (1.7.2001―30.6.2004; M. Liović) [SICRIS]
J1―3438: Lanosterol 14alfa-demethylase in biosynthesis of cholesterol and signalling sterols (1.7.2001―30.6.2004; D. Rozman) [SICRIS]
EC Framework 6 Grant (FP6-2002-LIFESCIHEALTH – LSH-2002-2.1.3-2 Eating disorders, from genes to behaviour) QLK1-1999-00916 Factors in Healthy Eating: The Role of Social, Genetic and Environmental Factors in Healthy Eating – a multicentre analysis of eating disorders and obesity (coordinators D. Collier and J. Treasure, U.K.); 2000-2003. R. Komel: Slovenian partner (MCMB: Medical Centre for Molecular Biology, Faculty of Medicine, Ljubljana, Slovenia). [Eating Disorders]
J3―2414: Genetic factors in eating disorders (1.1.2000―30.6.2002; R. Komel) [SICRIS]
J1―1350: Role of lanosterol 14alpha- demethylase in biosynthesis of cholesterol and strols with signalling properties (1.1.1999―30.6.2001; D. Rozman) [SICRIS]
CRP / SLO98-02 Molecular Genetic Aspects of Malignant Growth – ICGEB Collaborative Research Project (CRP, Contract No. 98/027), International Centre for Genetic Engineering and Biotechnology (ICGEB), Trieste, Italy & Medical Centre for Molecular Biology, Faculty of Medicine UL, Ljubljana, Slovenia, 1998-2001. R. Komel: Project Leader. [ICGEB CRPs Awarded]
J1―8762: Molecular genetics in biomedical research (1.1.1997―31.12.1999; R. Komel) [SICRIS]
INCO ECCACF European Community Action for Cystic Fibrosis [Commission of the European Communities, Joint European Project]. R. Komel: Slovenian Project Partner, 1996-1999. [SLO CF Mutation Spectrum]
Z1―7225: Human gene CYP51: determination of structure and regulation of expression (1.1.1995―30.6.1998; D. Rozman) [SICRIS]
ALIS 984/2-6 Biochemistry and Molecular Biology of Steroid-inducible Cytochromes P450. [Bilateral British-Slovenian Project ALIS 984/2-6, The British Council]: Krebs Institute, University of Sheffield, U.K. (Prof. Steven L. Kelly) & Institute of Biochemistry, Faculty of Medicine UL, Slovenia (Prof. Radovan Komel). R. Komel: Slovenian Project Leader, 1993-1994 and 1995-1996/7. [ ]
CFGAC The Cystic Fibrosis Genetic Analysis Consortium [Multilateral project for CFTR gene mutation analysis; coordinator: Lap-Chee Tsui, Toronto, Canada]. R. Komel: Slovenian Consortium & Project Member; 1990-1997. [SICRIS]
J4-7456: Molecular genetics in biotechnology oriented research on lower eucaryotes (1996 – 1997; R. Komel & V. Gaberc Porekar) [ SICRIS]
JEP 04297-94 Integration of Modern Cytogenetic and Molecular Genetic Methods in Education, and their Application [Commission of the European Communities, Joint European Project Grant S_JEP 04297-94, TEMPUS]. R. Komel: YU Project Leader, 1993-1994. [TEMPUS p.308]
Investigation of possible preparation of corticoid hormones by recombinant DNA methodology and application of the methodology in medicine. [Federal Project »Biotechnology of the Future«: Matić Fundation/Science Council of Yugoslavia]. R. Komel: Project Leader; F. Gubenšek (IJS, Ljubljana): Project Coordinator, 1988-1992. [ ]
F1-0509: Molecular biology in biotechnology and medicine: (C) Research on recombinant DNA methods for the synthesis ob biologically active compounds (1986 – 1991; R. Komel) [ ]
C1-0509: Molecular biology in biotechnology and medicine: (B) Molecular cloning at bioconversions of steroids (1986 – 1991 – 1992; R. Komel) [ ]
C2-0148/0504: Biosynthesis of lytic enzymes for fungal cel-wall degradation (1986 – 1992; R. Komel) [ ]
04-1677: Immobilization of microorganisms and biochemical investigations for application in biotechnology (1986 – 1991; R. Komel) [ ]
1.5.1.8.: Introduction of recombinant DNA methodology in diagnostic of selected diseases. [Federal Project GIBIT (Genetic Engineering & Biotechnology) / SZNJ (Science Council of Yugoslavia)]. R. Komel: Project Leader; Z. Kniewald (PBT, Zagreb): Project Coordinator; 1988-1990.




